tools for experimental design:
Command: randomizer, selectitems
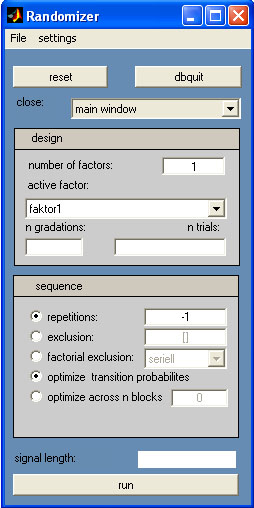
The randomizer is a tool for
generating pseudorandomized sequences, often required in EEG and MEG
experimental design, that relies on a large number of trials for
event-related averaging. Most often, the sequence must not be
predictable by the subject, to avoid expectancy effects.
Non-predictable condition sequences have equal transition probabilities
from one condition to any other, and the randomizer-program helps you
create such sequencies, by creating a large number of these sequencies
and selecting the best for your experiment. Sequence vectors are
displayed and saved in Presentation-Control-Language format to be used
with Presentation Software.
First, enter the number of
factors of your experimental design and hit return - the active factor popupmenu
then contains as many entries, one for each factor. For instance,
consider a 3 X 2 - design with the following number of trials per cell
|
|
|
factor 1
|
|
|
|
|
1
|
2
|
|
|
|
|
|
factor
2
|
1
|
|
30
Trials
|
30
Trials |
|
2
|
|
30
Trials |
30
Trials |
|
3
|
|
30
Trials |
30
Trials |
First, select faktor1 as active and enter the number of gradations. Hit
return and note the change of the entry for the active factor: faktor1 with 2 gradations becomes faktor1_2. Then enter the number
of trials for each gradation as vector (e.g. 90 90), and hit
return. Note again the change of the factor entries in the popupmenu: faktor1_23 becomes faktor1_2*90 90.Then select the
second factor as active and enter 3 gradations and 60 60 60 as
Trialvector (faktor2_3*60 60 60).
The total number of trials must be equal for all factors and is
displayed as signal length on the bottom of the randomizer window.
The resulting sequence for this setup will be a vector of 180 numbers,
with 30 ones, 30 twos, 30 threes, 30 fours, 30 fives and 30 sixes in
random order.
You can select a number of further restrictions concerning the order
of the condition numbers in the sequence:
The repititions-box allows you
to enter a maximum number of allowed repetitions of the same condition
in the sequence: entering a 3 will result in a sequence where the
same condition is allowed to occur at maximum 3 times in a row.
Use -1 to specify no
limits on repitions and deactivate the radiobutton or enter 0 to
exclude any repition.
The exclusion-box allows you
to specify condition number, that are not allowed to be adjacent to
each other. For instance entering 1 4 will result in a squence, where
each 4 is preceded and followed by any number but a 1, and each 1 is
preceded or followed by any number but a 4.
The factor exclusion-radiobuttonallows
you to avoid repetitions of a factor gradation: if activated the
sequence will not contain adjacent condition numbers, that share a
gradation of the same factor.
The optimize transition probabilites-radiobutton
decides how the sequence is generated: if inactive, only one sequence
is generated and displayed. For this sequence, the repitition and
exclusion limits are taken into account, but no further optimization is
performed. If active however, the generated sequence is optimized for
equal transition probabilites within the limitations imposed by the
repitition and exclusion parameters. This is done by generating a large
number of sequences to obtain an estimation for the expected transition
probability matrix of a randomly generated sequence. Each sequence is
then scored by calculating the "distance" (as sum of elementwise
absolute differences) from this expected transition probability matrix,
and a low distancefrom the expected matrix will indicate a well
balanced sequence. The distances are displayed as a frequency histogram
and will often be normally distributed.
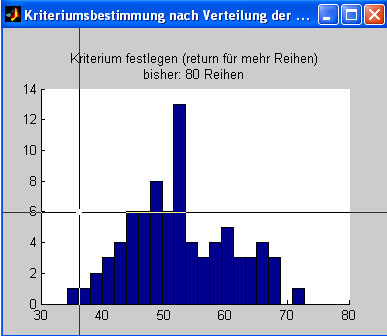
Click on the left part of the histogram to specify a maximum allowed
distance. The next sequence that will score below this threshold is
selected and displayed.
The result if saved by default to a textfile. A sample output is shown
below:
Randomizer:
Reihe mit 180
Elementen bestehend aus 6 verschiedenen Zahlen:
(mit
beliebiger Wiederholung)
array <int>
reihe[180]=
{1,1,5,1,6,6,3,1,1,6,4,1,2,4,3,1,6,6,4,2,1,1,5,3,6,4,2,5,2,2,6,5,4,2,3,6,5,6,2,1,4,5,1,3,1,1,4,5,2,5,6,3,1,6,3,3,3,1,4,2,2,4,4,2,1,4,6,3,2,2,2,2,6,5,6,2,2,6,6,3,6,4,6,5,6,4,5,2,5,6,4,2,3,6,1,3,5,4,3,5,2,3,2,6,5,5,5,2,1,3,2,1,3,4,5,1,4,3,3,3,4,4,3,5,4,1,5,5,3,1,4,2,6,6,6,5,2,5,3,5,3,5,3,2,3,6,1,5,4,4,6,5,4,4,2,4,3,4,4,3,6,2,1,1,1,2,4,5,1,5,1,1,4,6,1,3,3,2,5,6};
Übergangswahrscheinlichkeiten:
7
2 5 7
5 4
6
6 4 4
5 5
6
5 5 3
5 6
2
8 6 5
5 4
5
6 5 5
3 6
3
3 5 6
7 5

